Advanced Seminar
0368-4465-01
Lecture: Wednesday, 18:00-20:00, Schreiber 309
Notice a change in the schedule below (D.H., 19.12.04)
Instructors:
- Dan Halperin, [email protected]
- Nir Ben-Tal, Department of Biochemistry
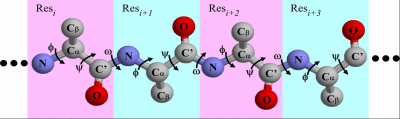
Kinematics is the study of motion of objects regardless of the causes of this motion. (Not to be confused with Kinetics.) There are close connections between the study of motion in robotics and in molecular biology. However, structural bioinformatics raises new and challenging computational kinematic problems due to the large
number of degrees of freedom typically involved. The goal of the seminar is to get the audience acquainted with kinematic issues (problems and some solutions) in structural bioinformatics. We will review techniques (heuristics as well as exact methods) and applications.
The seminar is primarily geared towards students with significant computational background. (It does not assume prior knowledge in molecular biology, but requires willingness to pick up the basics and relevant terminology.) This said, the first part of the seminar (inverse kinematics, loop closure) relies mainly on not-too-complex math. The second and shorter part (dynamic maintenance of kinematic structures) assumes familiarity with data structures and algorithms at the senior(last-year)-undergrad level.
The seminar will start with two introductory meetings, followed in subsequent weeks by student presentations. Here’s the program, revised on 10/11/04.
- 20.10.04 Introduction I (D.H.)
Kinematics: Brief introduction and basic terminology
Survey of the seminar’s topics and papers. - 27.10.04 + half of the meeting on 3.11 Introduction II (N.B.)
Introduction to protein structure
PART I: Inverse kinematics, loop closure, and related problems
- 03.11.04 + 10.11 (Amit)
- The Denavit-Hartenberg model, 3D direct and inverse kinematics
J. Craig, Introduction to Robotics: Mechanics and Control, Chapters 3 and 4
- The Denavit-Hartenberg model, 3D direct and inverse kinematics
- 17.11.04 (Idan)
- Cyclic coordinate descent: A robotics algorithm for protein loop closure
Canutescu A.A., Dunbrack R.L. Jr., Protein Sci. 2003 May;12(5):963–72 - A Randomized Kinematics-based Approach to Pharmacophore-Constrained Conformational Search and Database Screening
LaValle, S. M., Finn, P. W., Kavraki, L. E., and Latombe, J.-C., Journal of Computational Chemistry, 21(9):731–747, 2000
Slides [ppt]
- Cyclic coordinate descent: A robotics algorithm for protein loop closure
- 24.11.04 (Keren)
- A kinematic view of loop closure
E. A. Coutsias, C. Seok, M. P. Jacobson and K. E. Dill, Journal Computational Chemistry 25 (2004) 510–528
Slides [ppt]
- A kinematic view of loop closure
- 01.12.04 (Rafi)
- Geometric algorithms for the conformational analysis of long protein loops
J. Cortes, T, Simeon, M. Renaud-Simeon and V. Tran, Journal Computational Chemistry 25 (2004) 956–967 - Computing protein structures from electron density maps: The missing loop problem
I. Lotan, H van den Bedem, A. M. Deacon and J.-C Latombe, In Proceedings of WAFR 2004, 153–168
- Geometric algorithms for the conformational analysis of long protein loops
- 08.12.04 (Eitan)
- Assigning transmembrane segments to helics in intermediate-resolution structures
A. Enosh, S.J. Fleishma, N. Ben-Tal and D. Halperin, Bioinformatics, Vol. 20, Suppl. 1 (ISMB 2004), pp. 122–129
Slides [ppt]
- Assigning transmembrane segments to helics in intermediate-resolution structures
- 15.12.04 (Mira)
- Database of Macromolecular Movements
M. Gerstein, W. Krebs, and others [link] - Modeling macromolecular machines using rigid-cluster networks
M. Kim, G. Chirikjian, In Proceedings of (WAFR) 2004, 123–136.
- Database of Macromolecular Movements
PART II: Dynamic maintenance of large kinematic structure
- 22.12.04 (Iftah)
- Dynamic maintenance of kinematic structures, Algorithms for Robotic Motion and manipulation
D. Halperin, J.-C. Latombe, and R. Motwani, In Proceedings of (WAFR) J.-P. Laumond and M. Overmars (editors), A.K. Peters, Wellesley, 1997, 155–170
Slides [ppt]
- Dynamic maintenance of kinematic structures, Algorithms for Robotic Motion and manipulation
- 29.12.04 No meeting
- 5.1.05 (Yaron)
- Algorithm and data structures for efficient energy maintenance during Monte Carlo simulation of proteins
I. Lotan, F. Schwarzer, D. Halperin and J.-C. Latombe, Journal of Computational Biology, 11(2004), 902–932
- Algorithm and data structures for efficient energy maintenance during Monte Carlo simulation of proteins
- 12.1.05 (Efrat)
- Deformable spanners and applications
J. Gao, L. Guibas and A. Nguyen, In Proceedings ACM Symposium on Computational Geometry 2004, 190–199
- Deformable spanners and applications
- 19.1.05 (Eran)
- Dynamic maintenance of molecular surfaces under conformational changes
E. Eyal and D. Halperin, Manuscript, 2004 [pdf]
- Dynamic maintenance of molecular surfaces under conformational changes
