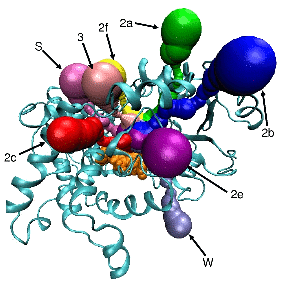
as detected by MolAxis. The CYP3A4 is represented by cartoons and the heme prosthetic group is represented by its VDW surface and colored orange. Each channel surface is colored in a different color for the sake of clarity.
Given a molecule modeled by a collection of balls in three-dimensional space we wish to efficiently identify pathways in the complement of their union. The desired pathways should balance between length and clearance. Namely, we prefer short and wide pathways between a given start point and goal point positioned in the complement. MolAxis is a web server and stand-alone software package designed to assist the biologist/biochemist to automatically identify pathways in the complement of molecules. A major contribution of the work is the notion of the pathway graph which approximates an idealistic construct related to the medial axis, and which unlike the medial axis is easy to compute in the case of the complement of the union of balls. We provide theoretical analysis of the properties of the pathway graph.
